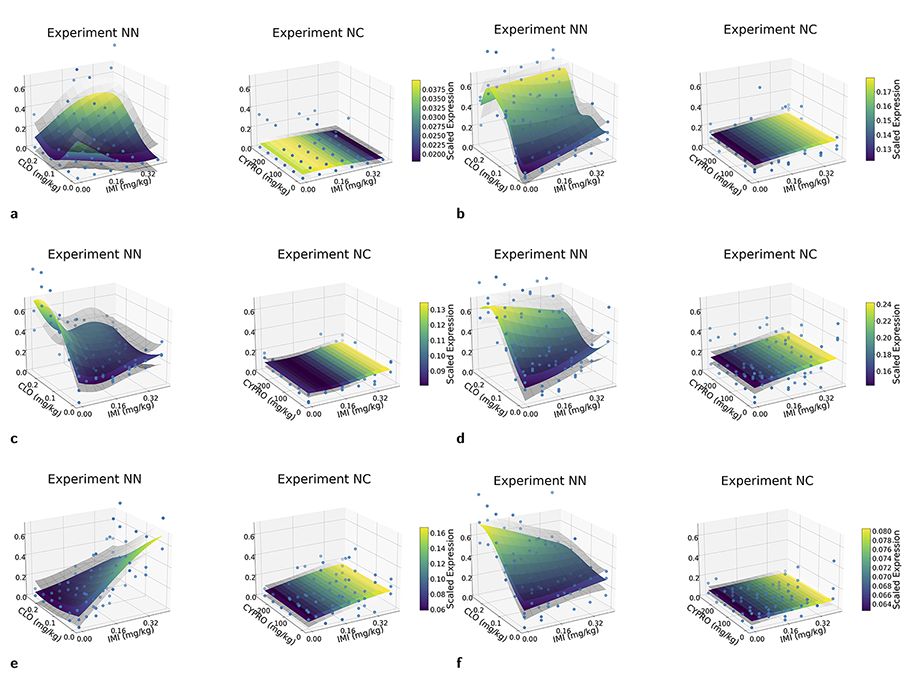
Gaussian processes for the analysis of mixture toxicity data
In the context of gene expression, mixture toxicity omics data involves analyzing how exposure to multiple chemical compounds affects the expression levels of genes. By studying gene expression profiles through transcriptomics data (measuring mRNA levels, i.e. gene expression), researchers can observe how various genes are upregulated or downregulated in response to chemical mixtures. This approach helps identify potential chemical interactions and how these effects differ from exposure to individual compounds. It is a key tool for understanding complex environmental exposures and their biological consequences.
In this research project, we will develop Gaussian process models for the analysis of mixture toxicity data. The data will involve 2-dimensional inputs corresponding to combinations of different concentrations of chemical compounds, and the response variable will be gene expression. We will extend two modelling approaches based on Gaussian processes. First, we will base the clustering framework GPclust [1] on it and extend it to 2-dimensional inputs. Second, we will extend our existing modeling approach to detect different types of interaction effects (additive, multiplicative) by incorporating negative binomial likelihood [2]. This will allow us to robustly detect differentially expressed genes even when the data are overdispersed/more noisy.
This project aims to extend existing computational methods and tools to allow the analysis of more complex biological data sets. If succesful, application to other, similarly complex datasets could be explored to evaluate the robustness of the methodology.
References
[1] https://github.com/SheffieldML/GPclust
[2] BinTayyash, Nuha, et al. "Non-parametric modelling of temporal and spatial counts data from RNA-seq experiments." Bioinformatics 37.21 (2021): 3788-3795.
Contact: Yuliya Shapovalova (RU); Dick de Ridder (RU, WUR).
Figure: Examples of expression patterns in differential gene expression analysis for two mixture toxicity experiments: 1) neonicotinoid (NN) and 2 ) cyproconazole (NC) exposures.